2016: The Loman Lab year in Tweets
31 Dec 2016January
“The past is a foreign country” – well, that’s how I feel about January 2016 looking back today. Definitely some things happened in January, but can’t remember them. So I’m using Twitter Analytics to remind me.
Oh! This was the month that #researchparasites came out, to the horror and amusement of the genomics field:
The logical fallacy of #researchparasites: expert data gatherers are highly unlikely to be the best people to analyse their own data.
— Nick Loman (@pathogenomenick) January 22, 2016
February
Was a good month. Our paper on the Ebola real-time genomic surveillance work came out, and it looked like the Ebola epidemic was well and truly over.
It's out! Real-time, portable genome sequencing for Ebola surveillance. https://t.co/fWkXHBpchZ
— Nick Loman (@pathogenomenick) February 3, 2016
Amazing work from @Scalene and many others
Behind the paper blog post on the Ebola surveillance. Warning: contains drama! https://t.co/RW1KDeiqoD
— Nick Loman (@pathogenomenick) February 3, 2016
There was also fun to be had at AGBT.
March
Just as we thought we had left Ebola behind, there was a flare-up in Guinea that spread to Liberia.
Samples from the new Ebola cases in East Guinea received yesterday; extracted & amplified overnight, sequenced today. Results inbound to WHO
— Nick Loman (@pathogenomenick) March 20, 2016
WHO update on Ebola flareup -we helped resolve source of cases with in-country sequencing: https://t.co/K7kCGkHvwV pic.twitter.com/P3yf2ZsFXj
— Nick Loman (@pathogenomenick) March 23, 2016
Phylogenetic analysis showed that the new cases were very closely related to an Ebola genome sequenced 500 days previously, as can be seen from this NextStrain tree.
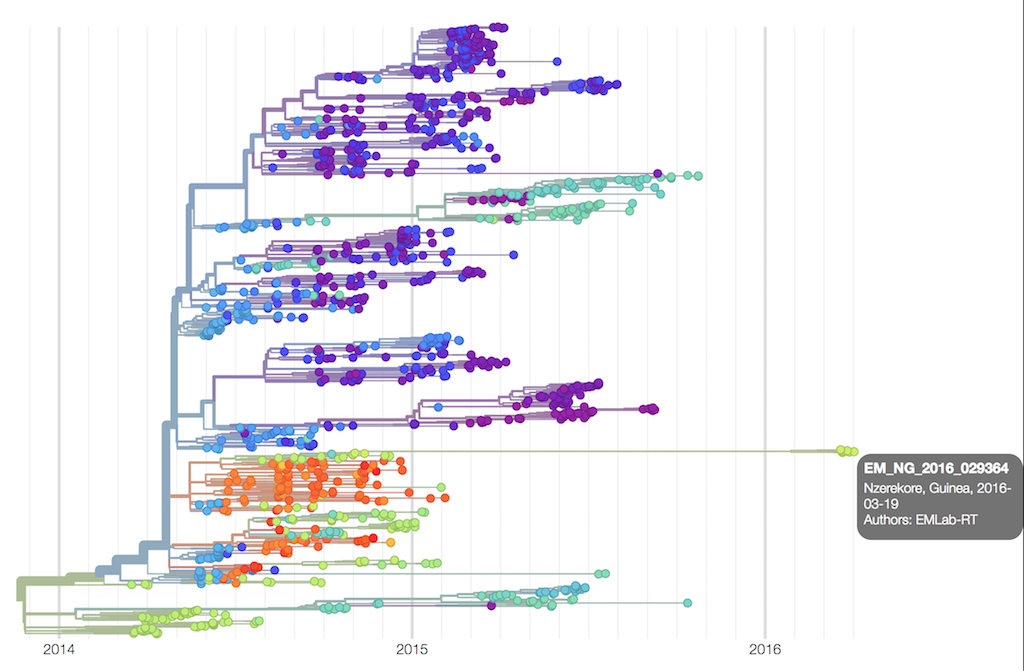
Independently, the epidemiologists identified a survivor who had been infected some 500 days previously, the very same individual.
This was a remarkable demonstration of the power of genomics, working in synergy with the epidemiologists on the ground.
Around the same time, we learnt we had been funded by MRC/Wellcome Trust/Newton to receive funds as part of the emergency response to Zika. Remarkably, the outcome was known just a few weeks after submitting the application, and we had the money just days after that. If only all grant funding could be like this …
Thanks MRC for funding our Zika seq lab in a caravan project w@nmrfaria @EvolveDotZoo @arambaut @edwardcholmes et al https://t.co/HXlEyLeNDX
— Nick Loman (@pathogenomenick) March 21, 2016
April
We wasted no time getting started. Josh flew to Sao Paulo to Ester Sabino’s laboratory to start testing out sequencing protocols for Zika.
That @scalene is a class act. 15 hour flight and 3 hours in customs last night and this evening he's sending over some Zika sequences.
— Nick Loman (@pathogenomenick) April 22, 2016
May
Oxford Nanopore released the R9 pore and it was something of a relief to see it was working well:
OMG, nanopore R9 just got a bit more real - first 2D read I BLASTed pic.twitter.com/p0493mL7s6
— Nick Loman (@pathogenomenick) May 17, 2016
Nanopore R9 does seem to give a nice shift in quality compared with R7.3, and with the added bonus of 250bp/s speed pic.twitter.com/JKWD3NBCVd
— Nick Loman (@pathogenomenick) May 8, 2016
We launched the ZiBRA project, a road trip around North-East Brazil to investigate the genomic epidemiology of Zika cases in this region, the most heavily hit by cases of microcephaly in newborns..
Today we launched the ZiBRA real-time viral genome sequencing project, follow along at https://t.co/ZpKp9x0EP4 @The_MRC @MRCeVal
— Nick Loman (@pathogenomenick) May 26, 2016
June
We hit the road for the ZiBRA project and started generating Zika genomes working in collaboration with the local public health laboratories. Lots of diaries and blog posts are on the Zibra project website if you want to read more about this trip.

We made lots of new lifelong friends in Brazil, and we didn’t die even though our bus caught on fire at one point, although we hit some technical obstacles with sequencing very very low abundance samples.
The year seemed to be going pretty well. Until the Brexit vote …
Going to bed feeling quite relaxed that the outcome will be fairly strong remain vote tomorrow. Don't make a mockery of this tweet overnight
— Nick Loman (@pathogenomenick) June 23, 2016
@pathogenomenick don't wake up!
— Neil Hall (@neilhall_uk) June 24, 2016
Not good.
I'm so sad about brexit there was a bit just then when I started singing along to Coldplay. Snapped out of it now.
— Nick Loman (@pathogenomenick) June 26, 2016
July
We launched the CLIMB cyberinfrastructure for microbial genomics to the public. Sign up for your own CLIMB account at the Bryn website. There are videos from the launch available, including this CLIMB demo.
Our preprint describing @MRCClimb, an amazing virtual compute infrastructure for microbial bioinformatics is now up! https://t.co/p5fH5TOu8u
— Nick Loman (@pathogenomenick) July 19, 2016
We've been working on this for a couple of years, finally (nearly) ready for the UK microbiology community! https://t.co/hITtJjOD6w
— Nick Loman (@pathogenomenick) July 13, 2016
The @MRCClimb architecture: https://t.co/p5fH5TOu8u Get started https://t.co/lagpSEdt9Z docs https://t.co/dIwMf38Gua pic.twitter.com/mBaAGYxhxs
— Nick Loman (@pathogenomenick) July 19, 2016
So far over 150 research groups in the UK have signed up for our virtual machine infrastructure which runs across three sites (Birmingham, Warwick and Cardiff) with Swansea to launch in 2017. Particular props to Radoslaw Poplawski, Tom Connor, Andy Smith, Marius Bakke and Matt Bull on the technical side who helped get this launched - just in time!
August
We sweated over the Zika sequencing protocol and eventually by the end of summer Josh Quick nailed something that worked well on samples with very low viral copy numbers.
In August we just about had time to fit in a week in Cornwall to teach Porecamp with Konrad and his crew.
Pablo, Emily, Jennie and Andy really got MicrobesNG motoring (over 7500 genomes sequenced with a median wait time of 6 weeks!), with insert sizes bolstered with a nice new Nextera XT protocol.
Kudos to @masonry_stoves, @bioemformatics and @scalene for developing this optimised Nextera XT protocol https://t.co/MthwWhz0Zw #gs2016
— Nick Loman (@pathogenomenick) August 31, 2016
Sep
The manuscript describing March’s Ebola flare-up was published.
Our study on a remarkable case from April 16 Ebola flareup; transmission from survivor *500* days post-infection https://t.co/TNk77RFoqS
— Nick Loman (@pathogenomenick) September 1, 2016
Zika genomes were coming out thick and fast thanks to the new protocol, our Brazilian collaborators, Sarah Hill and Alli Black. Josh’s third trip in 2017. A picture of Zika diversity was now starting to be built (beautifully visualised by Trevor and Richard’s wonderful Nextstrain site - vote for them to win the Open Science prize!).
Well done @scalene, @alliblk and Sarah Hill - smashing out the Zika genomes in Salvador, Brazil with @zibraproject since Friday
— Nick Loman (@pathogenomenick) September 11, 2016
A stunning new preprint from Andrew Rambaut, Gytis Dudas and the whole cast of Ebola sequencing collaborators was posted:
An unbelievably comprehensive phylogeographic study of 1600 Ebola genomes from @arambaut, @EvoGytis and others. https://t.co/3ywVp6XgKd
— Nick Loman (@pathogenomenick) September 2, 2016
We only managed one Balti and Bioinformatics in 2016 but it was a good one:
Ooh, it's #BaltiandBioinformatics today, featuring @koadman, @lexnederbragt and @torstenseemann ! I'll try and stream it on Hangouts.
— Nick Loman (@pathogenomenick) September 28, 2016
Christiane and the crew put on a good show at Genome Science.
Oct
I met Bill Gates and Nathan Myrvhold and gave a presentation at a “learning session” for Bill about using NGS to fight infection: it was incredible.
Life achievement unlocked. pic.twitter.com/6Y9v6sdLVv
— Nick Loman (@pathogenomenick) October 6, 2016
All the depressing news on Twitter got too much for me, and I took a break for 3 weeks. After 24 hours of extreme withdrawal symptoms, it was actually quite nice to do something else with my time, like imagining what people were saying on Twitter.
Relentless Brexit and Trump news is turning Twitter into an ordeal rather than a pleasure these days. (I realise this isn't helping).
— Nick Loman (@pathogenomenick) October 15, 2016
I've decided to take a Twitter/SM holiday until after the US general election. See you on the 9th Nov. Don't do anything stupid.
— Nick Loman (@pathogenomenick) October 17, 2016
@pathogenomenick you won't last a day
— Zamin Iqbal (@ZaminIqbal) October 17, 2016
They did, though.
(By the way Zam you were wrong).
Nov
Donald Trump - PEOTUS.
Wow what a terrible dream.
— Nick Loman (@pathogenomenick) November 9, 2016
Not a dream though.
Dec
A relaxed end to the year – we did a bit of Beach (well, Leith) sequencing with Andrew Rambaut and Tom Little in Edinburgh, look out for more BeachSeq action early in 2017..
And we even managed to release data from a 30x human genome on MinION working collaboratively on the sequencing with Nottingham, UCSC, UBC and Norwich, a mere 39 flowcells for that (assembly N50 - 3Mb!):
Initial FASTQ release of human NA12878 @nanopore whole-genome dataset available here (29 flowcells!) https://t.co/ZkirbOfztf#nanoporeconf
— Nick Loman (@pathogenomenick) December 1, 2016
~30X NA12878 nanopore data now posted up: https://t.co/Ln8mU44Y5Y - keen people might want to check out new rapid kit very long reads.
— Nick Loman (@pathogenomenick) December 16, 2016
Nanopore got named one of Science’s 10 breakthroughs of the year and we got a little name check.
Thanks @sciencemagazine for referencing our work in their feature on @nanopore as Top 10 Breakthroughs of the Year! https://t.co/3s7jeMZ2E0
— Nick Loman (@pathogenomenick) December 22, 2016
Finally in December, we heard the great news that the Ebola ca suffit! trial reported 100% efficacy for the Ebola vaccine. Well done Stephan, Miles, Sophie and all the others who worked on this.
A few changes in 2017
The MicrobesNG team sees a change in 2017 - we are sad to say goodbye to Andy Smith - our database programmer on the MicrobesNG project. He’s done an amazing job building the MicrobesNG website, our LIMS, the CLIMB Bryn site, and even had time to help out with the Zibra Project database and the Primal Scheme site. We cannot be too annoyed that he only spent a year with us – he’s had the once in a lifetime opportunity to become a trainee pilot with Aer Lingus, a lifetime dream for Andy.
There have been some changes in Birmingham too – it’s been really nice to have Alan McNally join the IMI as a new Senior Lecturer. And we are really excited that Willem van Schaik is joining the IMI later in April, Brexit be damned!
Politically we are in uncharted territory, so we enter 2017 with some trepidation about what is to happen to the scientific environment, but we also hope that the awesome wins out.
Happy New Year to all friends and collaborators from the Loman Lab!