Thar she blows! Ultra long read method for nanopore sequencing
09 Mar 2017tl;dr version
- Ultra long reads (up to 882 kb and indeed higher) can be achieved on the Oxford Nanopore MinION using traditional DNA extraction techniques and minor changes to the library preparation protocol, without the need for size selection
- The protocol is available here; it involves a modified Sambrook phenol-chloroform extraction/purification, DNA QC, minimal pipetting steps, high-input to rapid kit and MinKNOW 1.4
- We have tested it on E. coli and human so far with good results; data is of course available
Ultra-long reads: background
What if you could sequence E. coli in just one read? This was the challenge I set Josh. And why can’t we do that, if nanopore sequencing really has no read length limit?
Well actually: we’re not quite there yet, but we did manage to sequence 1/6th of the whole genome in a single read last week. Here’s how we (well, he) did it. As usual we like to release our protocols openly and early to encourage the community to test and improve them. Please let us know about any tweaks you find helpful! The community seems very excited by this judging by my Twitter feed and email inbox, so we have rush released the protocol. The tweets have also inspired commentaries by Keith Robison and James Hadfield, thanks guys!
Not bad .. 92% genome coverage of E. coli, average depth of 3.8x. But from just _43_ reads. https://t.co/vDqaEJblP9 (gt350kb.fasta) pic.twitter.com/x66o0lXayz
— Nick Loman (@pathogenomenick) March 2, 2017
First … a bit of background and the importance of working with moles not mass. This line of thinking was triggered during the Zika sequencing project when we noticed our yields when sequencing amplicons was never as good as with genomic DNA. Why was that?
We decided a possible reason is that nanopore sequencing protocols are usually expressed in terms of starting mass (typically 1 microgram for the ligation protocols). But of course 1 microgram of 300 bp fragments is a lot more (>25x more) DNA fragments compared to 1 microgram of 8000 base fragments. By not factoring this in the library prep, likely we were not making an efficient library because the protocol has not been scaled up 25 times to account for this difference. It stands to reason that it’s the molarity that’s important when loading the flowcell rather than the total volume of DNA. If you could load some imaginary molecule of DNA with mass 1000ng (bear with me), the chances of that interacting with the pore is still quite low. More molecules means more potential interactions with the pore, meaning more potential yield.
We calculated the desired starting molarity as 0.2 pM based on the length assumptions in the ONT protocol (in practice you load about 40% less after losses from library construction). So by increasing the amount of barcodes and adaptors, as we do in our Zika protocol, we can compensate for this.
That solves the short read problem, but we started thinking about how it would work in the other direction. What if you wanted to get the longest reads possible, what would this mean in mass? The rather silly idea was — if you wanted to get reads sufficiently long to cover a whole bacterial chromosome in a single read, what would the starting DNA concentration need to be?
The math here is simple; you just need to scale the starting DNA by 500x. But this would mean starting with ~500ug of DNA into the library preparation!
500ug of DNA is… quite a lot. And practically there are several problems with this idea:
- you would need a lot of cells to start with (perhaps not such a problem with bacterial cultures but certainly restrictive for some applications)
- what volume do you elute in? DNA starts to get viscous and thick as concentrations increase, at some point you just won’t be able to pipette any more
- how do you deposit that much DNA into the flow cell?
So - we slightly scaled down our ambitions and decided that it could be practical to scale up the protocol 10-fold, which could still result in average 80 kb reads, a significant improvement to the 8kb typically seen with the standard protocol.
We’d already been using the Sambrook protocol (from the classic Molecular Cloning - over 173,000 citations!) for our human genome extractions, which reliably gives very high molecular weight DNA that can be recovered with a Shepherd’s crook fashioned from a glass rod). Previously Dominik Handler demonstrated that HMW extractions with careful pipetting could generate long reads with the rapid kit. So we did a new Sambrook extraction using an overnight culture of E. coli K-12 MG1655 and generated something that was very pure (260:280 of 2.0) and very high molecular weight (>60kb by TapeStation - the limit of the instrument). In fact the DNA is so long that you can’t really size it without employing a pulse-field gel electrophoresis setup. Sadly we don’t have a working one in the department, so infrequently are they used these days. So we were flying blind in terms of the true length of the fragments.
Scaling up the rapid kit was relatively straight-forward when dealing with inputs up to 2 ug. You get DNA at a concentration of 250 ng/ul then add the maximum 7.5 ul. However to get inputs of 10 ug it requires concentrations of 1 ug/ul where things start to get tricky. The library is so viscous loading beads start to clump together and it becomes harder to get the library through the SpotON port on the flowcell. Not satisfied with 10 ug either we pushed on towards 20 ug which required making a double volume library and adjusting the dilution downstream. We eventually settled on a protocol which could reliably give read N50’s over 100 Kb (i.e. half of the dataset in reads of 100 Kb of length or greater) with a tail stretching out to 500 Kb, or sometimes beyond…
The final piece of the puzzle was something we were aware of; the nanopore control software as of version 1.3 does periodic ‘global voltage flicks’ - meaning that the voltage is reversed across the flow cell every 10 minutes. The aim of this is to prevent strands or proteins blocking up the pores, by a rapid change of the direction of the ionic current. However, the problem with a 10 minute flicking interval is that it intrinsically limits the longest read on the system to 150kb (with 250 base/s chemistry) and 270kb (with 450 base/s chemistry). In MinKNOW 1.3 you could change the script parameters (stored in a YAML file) to remove this flick, but in MinKNOW 1.4 luckily it has been dispensed with entirely in favour of a much smarter system that dynamically unblocks individual pores on demand.
So … how does it look after all that’s been done?
We ran E. coli K-12 MG1655 on a standard FLO-MIN106 (R9.4) flowcell.
E. coli stats
Total bases: 5,014,576,373 (5Gb)
Number of reads: 150,604
N50: 63,747
Mean: 33,296.44
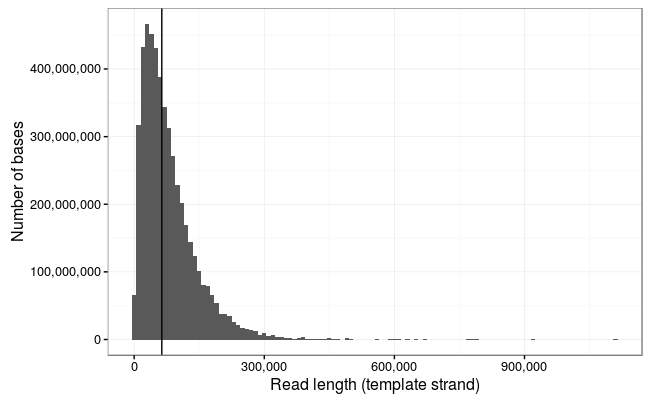
Read length stats
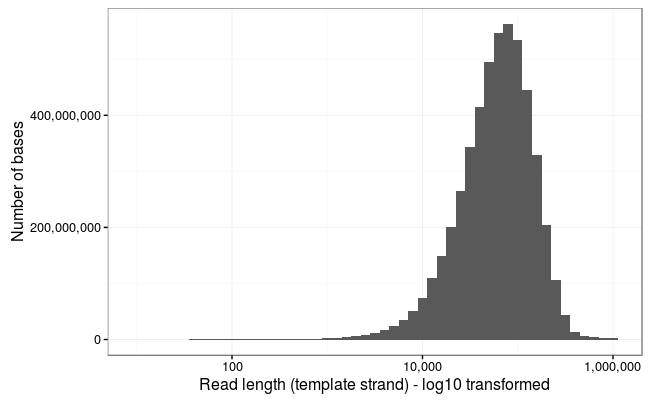
Ewan Birney suggested this would be more interpretable as a log10 scale, and by golly he was right!
Alignment stats
Wow! The longest 10 reads in this dataset are:
1113805 916705 790987 778219 771232 671130 646480 629747 614903 603565
!!!
But hold your horses. As Keith Robison likes to say, and Mark Akeson as well, it’s not a read unless it maps to a reference. Or as Sir Lord Alan Sugar might say, “squiggles are for vanity, basecalls are sanity, but alignments are reality”.
Are these reads actually real, then?
Just judging by the distribution it’s clear that this is not all spurious channel noise.
Let’s align all the reads…
Alignment issues
This dataset poses a few challenges for aligners. BWA-MEM works, but is incredibly slow. It goes much faster if you split the read into 50kb chunks (e.g. with split.py) but this is a bit annoying.
I decided to use GraphMap, this has a few useful functions - it will try to make an end-to-end alignment and it also has a circular alignment mode, which is useful as we would expect many of these reads would cross the origin of replication at position 0.
Another problem! The SAM format will not convert to BAM successfully, so I’ve output using the BLAST -m5 format for ease of parsing. The SAM/BAM developers are working on this (CRAM is fine).
After a solid couple of days of alignment, here are the results:
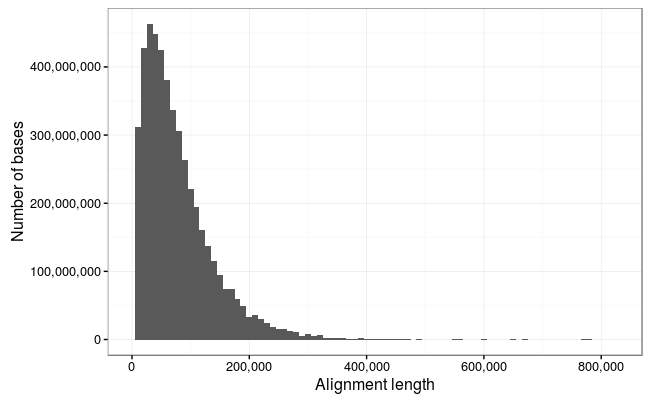
So we lose a few of the really long reads here which are obviously noise (the 1Mb reads is just repetitive sequence and probably represents something stuck in a pore and the 900Kb read is not a full-length alignment), but otherwise there is an excellent correlation between the reads and alignments.
So, the longest alignments in the dataset are:
778217 771227 671129 646399 603564 559415 553029 494330 487836 470664
That’s theoretical 1x coverage of the 4.6Mb chromosome of E. coli in just the 7 longest reads !!
95.47% of the bases in the dataset map to the reference, and the mean alignment length is slightly higher at 34.7kb.
A few other notable things:
- The 790kb read that didn’t align full-length is interesting. On inspection it is actually two reads - the template and complement strand of the same starting molecule, separated by an open pore signal. This gives us a clue as to how the proposed 1D^2 technology (which is replacing 2D reads) could work. Calling the two reads together (thanks, Chris Wright) gives a 95% accuracy read!
- We’ve started using the Albacore basecaller for this, rather than uploading to Metrichor. Albacore seems to keep up with basecalling a live-run when using 60 cores.
Summary
So we would like to claim at least four world records here!
Longest mappable DNA strand sequence **
Longest mappable DNA strand & complement sequence
Highest nanopore run N50 (not sure about other platforms?)
Highest nanopore run mean read length
(**) we’ve actually beaten that record already with another run, but a subject for another post
An interesting exercise for the reader is to figure out the minimum number of reads that can be taken from this dataset to produce a contiguous E. coli assembly! My first attempt found a set of 43 reads which covers 92% of the genome, but you can do better!
Where now? Well, readers will notice that a real landmark is in sight - the first megabase read. We’ve been running this protocol for a bit over a week and a new hobby is ‘whale spotting’ for the largest reads we can see.
We haven’t quite yet worked out a systematic naming scheme for whales, but perhaps Google has the answer.

So in that case, we’ve in the past few days hit our first narwhal (an 882kb read from a different run, which translates to a 950kb fragment judged against the reference).
How can we go longer? Well it might be possible to increase the DNA input some more, but we start hitting issues with the viscosity which may start to prevent pipetting onto the flowcell. Also pipette shearing forces are presumably an issue at these concentrations.
The general consensus is that we will need to employ solid-phase DNA extractions and library construction, e.g. in agarose plugs. The SageHLS instrument also looks quite interesting.
Data availability
Hosting courtesy of CLIMB:
- GraphMap alignment
- Basecalls (5Gb)
- FAST5 files (500Gb)
- Poredb database (Poredb for details on Poredb)
Acknowledgements
The nanopore squad, John Tyson and Matt Loose provided much helpful advice and input during the development of this protocol. Matt Loose came up with the whale naming scheme.
Thanks to ONT for technical support with particular thanks to Clive Brown, Chris Wright, David Stoddart and Graham Hall for advice and information.
Conflicts of interest
I have received an honorarium to speak at an Oxford Nanopore meeting, and travel and accommodation to attend London Calling 2015 and 2016. I have ongoing research collaborations with ONT although I am not financially compensated for this and hold no stocks, shares or options. ONT have supplied free-of-charge reagents as part of the MinION Access Programme and also generously supported our infectious disease surveillance projects with reagents.