Sample preparation and DNA extraction in the field for nanopore sequencing
13 Oct 2016The nanoporati are currently thrilling to a bevy of new announcements from Oxford Nanopore Technologies (ONT). More information over at the “wafer-thin update” and insightful commentary from Keith Robison on his blog.
But amongst the noise and excitement of future products, there are three important updates we are focusing on right now:
- the release of the 5-10 minute 1D rapid prep (Mu transposase based)
- coupled with the new R9 (now R9.4) chemistry that produces usable and high accuracy 1D reads (both discussed in this previous post)
- and, new updates to the pore, membrane, motor and loading protocol which suggest 5-10Gb output may now be achievable.
Bazinga! 10G. Next kit. pic.twitter.com/GhBN0usiH8
— Clive G. Brown (@Clive_G_Brown) October 5, 2016
We just received our first R9.4 double-speed (450 b/s) kits and so we will see how it looks soon, but as of now, we are able to get up to 3Gb of output on the vanilla R9.
The significance for our work: we can now start to consider using MinION for metagenomic sequencing (previously we have restricted our ambitions to sequencing individual viruses and bacterial cultures due to relatively low outputs).
Ultimately our research group would like to get to culture-free diagnosis of infectious diseases, with full genomic coverage, as a near-patient assay. There have been a few proof of principle papers here including use on Ebola and chikungunya (from Charles Chiu) and on bacterial urinary tract infections (from Justin O’Grady).
However, for portable metagenomics sequencing to really become a viable prospect, the sample needs to be rapidly prepared at point of collection (from near the patient, in diagnostics, or from water, food, animals, the natural environment, etc.).
In my view, local sample preparation, DNA extraction and local bioinformatics analysis are now the major open issues for portable sequencing.
To illustrate this point, we recently saw the exciting news that the nanopore had been run on the International Space Station - surely a landmark moment in genomics. But yet the sample was still not prepared or DNA extracted in space.
And sadly, you cannot get away without proper DNA extraction. We saw in the past few days the bizarre spectacle of David Eccles and Chris Mason at a conference in Australia attempting to sequence various food samples (coffee, strawberries and cream, etc.) on the nanopore, using the 1D prep. A valiant experiment, but the output was effectively noise due to a lack of pure DNA prep. We experienced similar results when attempting to sequence from a virtually DNA free sample on the beach in Cornwall.
So, DNA extraction remains a fact of life for sequencing.
For single molecule sequencing it’s even trickier: you need high purity, high molecular weight, high concentration DNA to get good results from single molecule sequencers like the nanopore (current input 500ng for the transposon prep).
This should not be problematic for many environmental samples. When dealing with low concentration samples, the easiest way of doing this is via PCR (targeted or untargeted WGA, although fragment length can suffer without significant optimisation).
Solutions for portable sample preparation
Whilst presumably not a big market yet, a few companies have started producing solutions for ‘in-field’ sample preparation and DNA extraction. In the rest of this post I want to explore some of the available options for portable sample preparation and DNA extraction.
Just as a reminder, the steps for sample preparation are to a) make the sample safe (particularly important e.g. in Ebola) b) homogenise the sample and lyse cells c) extract DNA and then d) make a sequencing library.
Microbiome maven Elizabeth Bik has a very nice review of this in a recent article which is focused around microbiome studies but applies equally to other types of study:
Bead Beating/Tissue Lysis
Many samples, and particularly environmental samples, need homogenisation and cellular disruption before DNA extraction can proceed efficiently. One of the most popular methods is bead beating, which usually requires a benchtop instrument. Luckily, there is a portable, battery-powered method available in the form of the TerraLyzer (we have one). This device available, available from Zymo Research, uses a converted power tool to act as a portable bead beater. It’s a solid bit of kit, but costs about $1000. If that’s too rich for you, Russell Neches has developed a template that can be 3D printed to turn a Craftsman automatic hammer into a portable bead beater.
Here is a video of Russell using the TerraLyzer to extract DNA from cat poo:
DNA extraction
DNA can be extracted very simply from a variety of foods like bananas or strawberries and is method probably familiar to school children; First fruit is blended to break up the tissues, washing up liquid is then added to breakdown the cell membranes before being straining to remove solids. DNA is then precipitated by adding alcohol and spooled off using a toothpick. More detailed instructions here: http://biology.about.com/od/biologylabhowtos/ht/dnafromabanana.htm.
100% ethanol is a problematic substance to ship (it is banned from aircrafts), so it is an open research question about whether a lower proof alcohol that is readily available, e.g. vodka, would be an acceptable substitute. Please fund this important project.
Portable devices
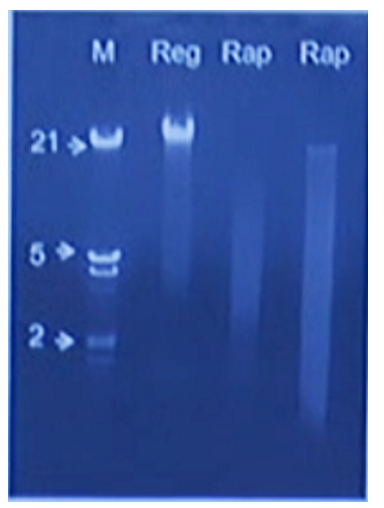
Claire Lonsdale brought along an interesting device to Porecamp in Cornwall called the PureLyse from Claremont Bio. This device combines bead-beating with DNA capture using silica beads which are agitated by a small motor. They have built a small disposable device which combines a syringe and a reusable battery pack. The sample, ideally bacterial culture, is aspirated via the syringe then the motor is turned on for a minute to burst the cells/bind the DNA. Claire presented results at London Calling demonstrating that the DNA extracted is probably suitable for PCR but may be too fragmented for single-molecule sequencing.
Screenshot from Claire’s London Calling talk, showing rapid extraction on the right compared to a regular spin-column extraction on the left.
The announced, but not currently available Zumbador from ONT looks to take this syringe concept further, by including reagents for lysis, purification and potentially library preparation in a single pre-loaded cartridge. This looks appealing but the worry for those of us who deal with a lot of DNA extractions from different organisms is which cell lysis solution is likely to be universally applied to all manner of organisms with quite different cell wall compositions - Gram positives and spore forming bacteria are notoriously tough shells to crack, this may need to be combined with the bead beating step above.
Zymo also offer the Xpedition kit range, which are designed with field work in mind and contain a stabilisation solution which will preserve your DNA (after bead beating with the TeraLyzer) for up to a month at room temperature.
However, you can also use traditional column-based extraction method in the field, if you have a:
Portable centrifuge
From my research, microcentrifuges are nearly all mains powered, which limits their utility in the field.
A homebrew solution is simply to modify a cordless drill with a 3D printed centrifuge adaptor, one example being the DremelFuge, that offers up to 52,000g/rcf acceleration.
However one should be extremely careful here because a flying, solid object at these rotations could cause serious harm, please take appropriate safety precautions if you are thinking of using this solution. Disclaimer! More generally, if in doubt about any safety aspects of field sample preparation, please first get in contact with your local safety officer for advice.
An alternative is to adapt a regular lab microcentrifuge that can take DC input, as that means they can be easily powered from a Lithium-Ion battery pack.
Portable PCR Thermocyclers
The MiniPCR is a fantastic (we’ve got one) biohacker/kickstarter product which costs £500 from Cambio in the UK. It is programmed via a laptop or phone but then must be plugged into a mains adapter or battery pack to start the program. We bought a LiPo powerbank off Amazon for £70 which can provide the 19V, 3.7A power requirement. They also produce a small electrophoresis and visualisation system to go with it.
An alternative is the Bento Lab from Bento Bio. This device caught many people’s attention with its Fisher-Price toy looks and intriguing functionality - it is a PCR thermocycler, gel visualiser block and minifuge all in one! Although mains powered, it should draw sufficiently little power that it could be powered via a car battery or possibly a Lithium pack. We had the pleasure of seeing a prototype box and it kicks ass - the only problem at the moment is that it’s still not available to buy. I hope it will ship soon and we’ll be first in the queue to test it out.
## Portable Liquid Handler
Pipetting is only accurate at relatively large volumes (>1 ul) which both increases reagent costs and can be a major source of errors with multi-step protocols. The Voltrax is an interesting device that was announced by ONT at London Calling 2015 and has not yet been seen in the wild, although the access programme was recently announced. The basic principle is the movement of ultra low liquid volumes around a matrix through an applied electrical current - a process called electrowetting. The appeal of such a process is that complex pipetting and mixing steps could be automated (apparently via a scriptable Python interface).
There may well be more that I have not mentioned … feel free to drop your suggestions in the comments box below!
Conflict of interests
I have received an honorarium to speak at an Oxford Nanopore meeting, and travel and accommodation to attend London Calling 2015 and 2016. I have ongoing research collaborations with ONT although I am not financially compensated for this and hold no stocks, shares or options. ONT have supplied free-of-charge reagents as part of the MinION Access Programme and also generously supported our infectious disease surveillance projects with reagents. Cambio sent us some free reagents to go with the MiniPCR instrument we purchased.
Credits
Thanks to Josh Quick for contributing to this post, and to Matt Loose and John Tyson for reading a draft version.